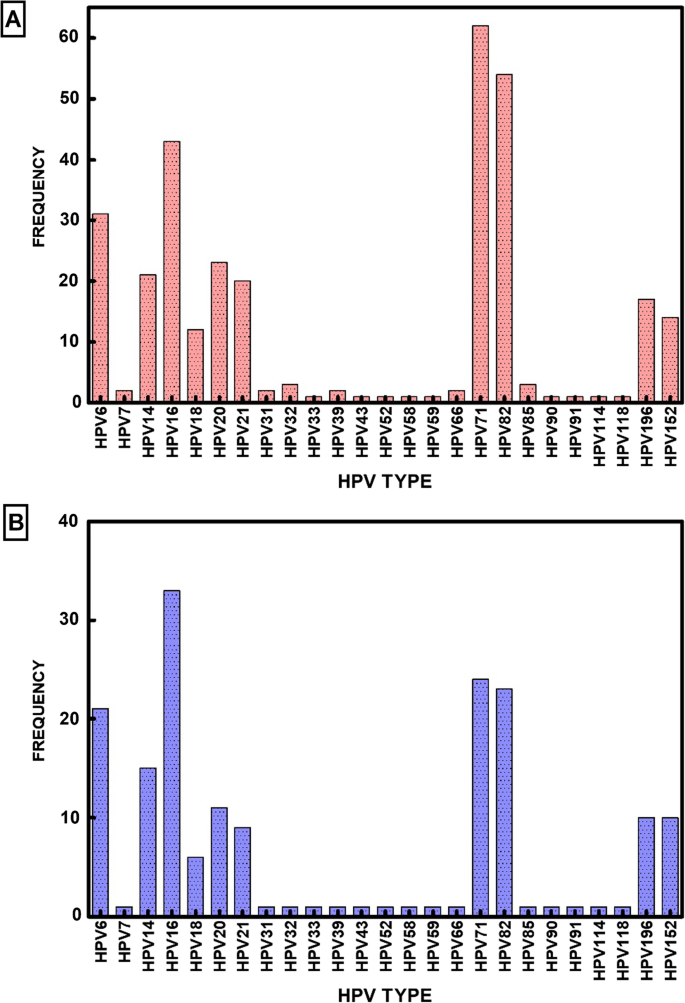
This study’s objective was to use DNA amplification and sequencing to ascertain the variety and prevalence of HPV types in Nigeria. Using a condensed FTA card protocol( GE Life Sciences, Piscataway, NJ ) methodology, the samples were gathered and transported. The DNA samples were amplified by RCA. NGS sequencing and tsPCR were used to analyze the amplified DNA. 90 cervical samples from two regions of Nigeria were used in our analysis; the results are shown in Additional file 2: Table S2. In these 90 samples, NGS sequencing and tsPCR revealed a diverse group of 25 HPV types( Fig. & nbsp, 1. HPV types 6, 7, 14, 18, 20, 21, 31, 32, 33, 43, 52, 58, and 69 are found in a critical analysis of the results. They are also found to be 91, 85, 90, 1, 114, 128 ( 152 ), and 6 ( 69 ). Major HPV types that are prevalent in this population should be taken into consideration( Table 1 ).
HPV genotype detection and identification using NGS
44 different HPV types were found in this cohort of cervical samples after NGS was used to identify them. The following HPVs were identified: 6, 7, 11, 14, 18, 21, 25, 31, 32, 33, 34, 43, 45, 52, 53, 61, 70, and 69, as well as the following: 71, 78, 82, 85, 886, 987, 90, 1, 114, 128; 195, 220; and 228. The most frequently detected Hr-HPV type was HPV82, and the most commonly detected Lr – HV types were HPS71 and HPH72. Due to its capacity to identify numerous HPV-type infections and novel types that may elude other molecular methods, this observation suggests that NGS is a highly sensitive method for HIV detection and typing. Overall, we discovered 18 Hr – HPV, 13 Lr, and 13 Ur, all of which indicate NGS’s unbiased detection strategy. Additionally, 10 of the 13 Ur-HPV types discovered were beta, 2 were gamma, and 1 was alpha. This is in line with the NGS’s capacity to identify numerous infections involving various papillomavirus families.
According to our findings, the distribution of HPV types was as follows: Ur-HPV, with Ur-40, Hr-34 %, and Lr-26 %, respectively( Table 2 ), was higher than Hh-HpV and R-VV. The most common strain is HPV type 16, followed by types 6 and 18, which are also somewhat common. However, NGS discovered a number of less well-known HPV types, including 14, 19, 20, 21, 25, 71, and 82, which seem to be common among Nigerians. In the cohort of 90 samples, we were unable to find any samples that were missing HPV DNA.
In the cohort, 80 samples contained multiple HPV type infections between 2 and 19 different types, whereas only ten samples had a single HIV type infection. Multiple HPV-type infections were more common overall at 87.78 %( 95 % CI 79.43-93.04 %). A startling result of the NGS analysis is that 31 out of 90 patients appeared to have four or more HPV types at once. Single HPV type infection, on the other hand, was low at 12.22 %( 95 % CI 6.96 – 20.57 %).
To calculate the prevalence of HPV risk types in both single – and multiple-infection infections, we calculated their frequency. Using NGS detection, we discovered that Lr – HPVs were higher in single infections with 57.55 % compared to 4<a data-track="click" data-track-label="link" data-track-action="table anchor" href="https://virologyj.biomedcentral.com/articles/10.1186/s12985-023-02106-y#Tab3“>3.45 %, <a data-track="click" data-track-label="link" data-track-action="table anchor" href="https://virologyj.biomedcentral.com/articles/10.1186/s12985-023-02106-y#Tab3“>3<a data-track="click" data-track-label="link" data-track-action="table anchor" href="https://virologyj.biomedcentral.com/articles/10.1186/s12985-023-02106-y#Tab3“>3.<a data-track="click" data-track-label="link" data-track-action="table anchor" href="https://virologyj.biomedcentral.com/articles/10.1186/s12985-023-02106-y#Tab3“>3<a data-track="click" data-track-label="link" data-track-action="table anchor" href="https://virologyj.biomedcentral.com/articles/10.1186/s12985-023-02106-y#Tab3“>3 %, and 66.67 %, respectively. We found that using nGS detection ( Table <a data-track="click" data-track-label="link" data-track-action="table anchor" href="https://virologyj.biomedcentral.com/articles/10.1186/s12985-023-02106-y#Tab3“>3 ) in samples with multiple infections, Ur – HPVs were detected at a higher rate of 44.21 % vs. <a data-track="click" data-track-label="link" data-track-action="table anchor" href="https://virologyj.biomedcentral.com/articles/10.1186/s12985-023-02106-y#Tab3“>32.05 % and 2<a data-track="click" data-track-label="link" data-track-action="table anchor" href="https://virologyj.biomedcentral.com/articles/10.1186/s12985-023-02106-y#Tab3“>3.74 %) than Hr-HPV. According to Table <a data-track="click" data-track-label="link" data-track-action="table anchor" href="https://virologyj.biomedcentral.com/articles/10.1186/s12985-023-02106-y#Tab3“>3, using NGS + ts – PCR combination, Lr-HPVs were 22.5 %, Hr-1. 88 %, and Ur-HV-<a data-track="click" data-track-label="link" data-track-action="table anchor" href="https://virologyj.biomedcentral.com/articles/10.1186/s12985-023-02106-y#Tab3“>35. 6<a data-track="click" data-track-label="link" data-track-action="table anchor" href="https://virologyj.biomedcentral.com/articles/10.1186/s12985-023-02106-y#Tab3“>3 %, respectively.
Type-specific PCR confirms the HPV genotypes found.
Due to the high sequence homology of HPV types, the wide variety of virus family members, and the possibility of multiple HIV strains infecting a single person, NGS analysis is arguably the most sensitive method for HIV analysis. Even for NGS, we could not ignore the possibility of false positives. As a result, we made the decision to use single ts – PCR analysis to examine each HPV type found by NGS in each sample. In order to eliminate any potential sequencing errors as well as any errors that might arise due to high sequence homology among various HPV types, the NGS data of these types were critically re-examined using a second methodology. using type-specific PCR ( ts-PCR ) for each of the 44 HPV types that NGS has identified. We confirmed 25 HPV types, including 7 Lr-HPV, 7 Ur-HV-, and 11 Hr – HV. In the sample cohort for this study, we discovered HPV types 3, 6, 7, 14, 18, 20, 21, 32, 33, 43, 52, 58, and 69, as well as 81, 85, 90, 91, 114, 128. HPV14, 16, 20, 71, and 82 were the five genotypes that were most frequently found among the types found in descending order.
to confirm the ts – PCR detection rate in order to determine the HPV infection risk types. We discovered that HPV16 was the Hr-HPV type that was most frequently detected, followed by HPHV71 as the most commonly detected Lr– HVP type, and HPP1<a data-track="click" data-track-label="link" data-track-action="table anchor" href="https://virologyj.biomedcentral.com/articles/10.1186/s12985-023-02106-y#Tab4″>4 as being the Ur-HVA type with the greatest number of detections. Ts – PCR( Table <a data-track="click" data-track-label="link" data-track-action="table anchor" href="https://virologyj.biomedcentral.com/articles/10.1186/s12985-023-02106-y#Tab4″>4 ) failed to identify 19 HPV types from NGS sequencing. Our findings suggest that six of the seven Ur-HPV types identified by ts-PCR were beta papillomavirus, and one was alpha. In Table 2, we found that the detection rate of NGS + ts- PCR for Hr-HPVs was <a data-track="click" data-track-label="link" data-track-action="table anchor" href="https://virologyj.biomedcentral.com/articles/10.1186/s12985-023-02106-y#Tab4″>40 %, followed by 27 % and 33 %, respectively.
The prevalence of single HPV-type infections was generally low. We determined the frequency of these types in order to determine the rate of HPV risk types found in infections of both single-type and multiple types. We discovered that the prevalence of HrHPVs was higher in single infections( 66.67 % vs. 33.3 %). There were no single infections where Ur – HPVs were found. Using NGS + ts – PCR combined detections ( Table 3 ) and a distribution of 41.88 %, 35.63 %, and 22.50 %, respectively, we discovered more HrHPVs in multiple infections compared to UrHPVPs or LrHVS.
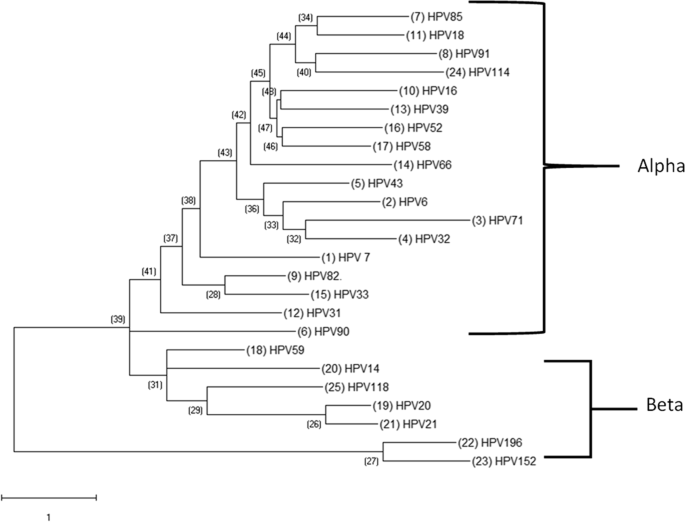
Phylogenetic analysis of HPV DNA
After examining the evolutionary patterns of a few HPV types, we were able to distinguish between two distinct clade groups: one group that included the papillomaviruses and the other group of the same viruses. Compared to the – clades, the”- clade” possessed the greatest evolutionary diversity. This suggests a significant evolutionary divergence between the papillomaviruses. In terms of the number of substitutions per site ( Fig. ), HPV71 also displayed a longer branch than the other papillomavirus genus members. & nbsp, 2.
Select Papillomavirus found in the cohort: Phylogenetic Analysis. Each papillomavirus’s HPV DNA sequence was obtained from PAVE and used for MEGA analysis. A phylogenetic tree was then created using the multiple alignments that resulted in order to examine the evolutionary patterns of a particular papillomavirus. The alpha – papillomavirus genera and Beta, which are two genera of the PV, were used to identify plaques that were connected to the classical Papillipomavius genera. The Maximum Likelihood method and the Kimura 2 – Parameter model from the MEGA package were used to create phylogenetic trees.